It’s been a while since I wrote a blog post on a new paper, but the recently published paper of Jill de Ron et al. investigates limitations of network models discussed for a good decade now, and I’m very happy it’s online (URL, PDF).
As many of you know, network models in our field often consist of observed variables, and the focus has often been on symptoms. For example, insomnia, sad mood, and concentration problems are depression symptoms. Single items (e.g. “how well did you sleep in the last 2 weeks”) are often used to assess symptoms (here: insomnia), because diagnoses often include a large number of symptoms, and asking multiple questions per symptom can add considerable burden to participants, especially in transdiagnostic contexts where researcher assess not one, but many diagnoses.
Because single items can be unreliable, psychometrics has a history of trying to capture latent variables with a number of items to remove item-specific (but construct-unspecific) measurement error. Psychometricians are interested in the shared variance of a number of items measuring sleep problems, rather than the unique variances of the specific items that may be due to odd phrasings or other item-specific peculiarities. This, in a nutshell, is why many questionnaires have more than one item. And also why we measure mathematical intelligence or personality facets or students’ exam performance with multiple questions, not just one.
There has been much speculation about the role of measurement error in network psychometrics, but thorough investigations have been absent.
Research questions
In the paper, we have four broad research questions.
First, we want to understand how much measurement error interferes with the estimation of cross-sectional network models. This can be done via simulation studies: we have a true network structure, simulate data (in this case, the 9 DSM-5 depression symptoms), and estimate back the network structure. In many scenarios, the true network will be recovered very well, which has been shown in various different papers over the years. Now, we do the same thing again, but we introduce some measurement error to the data, and then see how much worse the true network will be recovered. Rinse and repeat.
Second, we want to understand what situations are more problematic than others when it comes to measurement error. For example, would you rather want a small sample size (not great for recovering networks) and little measurement error, or a large sample size (much better for recovering networks) and moderate measurement error? A simulation study allows doing this by varying a number of parameters in the simulation. So in addition to varying the degree of measurement error in a simulation, we can also vary other parameters, such as sample size.
Third, when we have multiple indicators per symptoms, in what ways should we estimate network models to mitigate measurement error? To do so, we simulate data now, but instead of simulating 9 depression symptoms, we simulate 3 questionnaires with 9 DSM-5 depression symptoms that are somewhat different from each other (so each of the 3 questionnaires has 9 items, and that means that we can now use e.g. the 3 insomnia items from the 3 different questionnaires to estimate a latent variable for insomnia). We then can fit various models and see which models best recovers the true structure:
- Single indicator networks: 3 separate network models (M1a, M1b, M1c), based on the 9 symptoms from each scale (so we fit a network to questionnaire 1, then questionnaire 2, then questionnaire 3 separately, each network has 9 nodes).
- Multiple indicator networks: 3 separate network models that take into account measurement error in various ways. (M2) estimates average symptom scores across the 3 questionnaires (e.g. we average the 3 insomnia items from the 3 questionnaires into 1 insomnia item); (M3) estimates symptoms based on factor scores (we fit a factor model instead of averaging); and (M4) estimates latent network models, a statistical technique developed by Sacha Epskamp that combines both factor score estimation and network estimation in one step
Here is an example figure from our paper; on the left, you can see that we simulate data under a true structure where each of the 9 latent symptoms (round nodes) have 3 observed indicator symptoms each (quadratic nodes). E.g. a round node could be “insomnia”, and the 3 indicators could be 3 separate questions about sleep. On the right side, you then see various ways in which we estimate the network structure: using only one of the questionnaires, averaging the observed symptoms (e.g., the 3 insomnia questions), estimating a factor model and using those factor scores in a network model, or doing it together in a latent network model procedure (LNM).
(Click on the images in this blog post to zoom in)
Fourth, when we were working with the STARD data years ago, we realized they measured depression symptoms using 3 separate questionnaires. All of them cover the 9 DSM-5 depression symptoms, so we can now actually estimate all the models we mentioned above (e.g. one network per questionnaire, or combining the items in various ways using average symptom scores or factor models). Now, we of course don’t know which of these models get it “right” in the STARD data because we do not know the true model. But we can check for the agreement of these estimation methods. One would assume that a model that gets very different findings than all the other findings would be the least reliable method.
Results
I will not go through all results in detail … there are many. Instead, I will zoom in on some important findings.
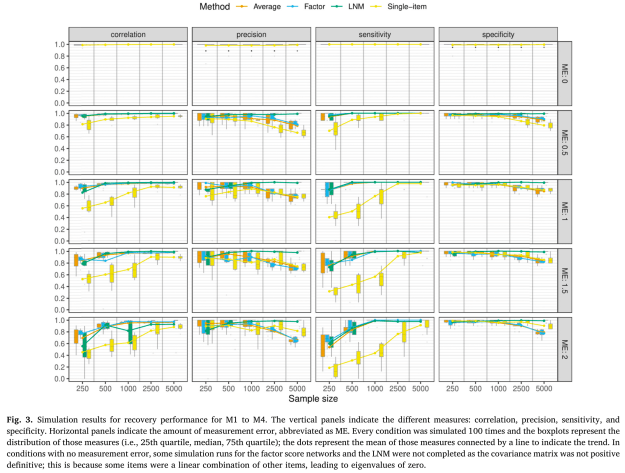
First, the results of the different estimation methods in recovering true structures under varying degree of sample size and measurement error. Broadly, you can see in the top row that estimation performance is perfect when there is no measurement error (ME: 0). Single indicator networks, even in relatively small samples of n=250, recover true structure very well.
Second, generally, the single-item networks in yellow perform worse than the other estimation methods across most conditions. When you estimate networks via single indicators, metrics such as the correlation between the estimated and true network structure (first column) perform worse (i.e. lower correlations) in single indicator networks compared to multiple indicator methods. Sensitivity (third column) is also lower for single indicator networks.
Third, the 3 multiple indicator methods perform quite similarly, including the very simple method of averaging across items; there are some differences, but they are quite small.
Fourth, we can see that the recovery performance, even under severe measurement error (ME: 2) is quite good when N reaches around 500 through 1000. This highlights, once more, and against popular claims in the literature, that recovery performance of basic network models in cross-sectional data can be excellent.
Lets now turn to the empirical STARD dataset, a dataset of around 3500 participants who will undergo treatment (we only utilize the baseline measurement point before treatment starts).
You can see here (first row) the estimated networks using the 3 separate questionnaires (QIDS-C, QIDS-S, IDS-C). We estimate models on the 9 DSM-5 symptoms, but do so in a disaggregated way, that is, we split up opposing symptoms (e.g. insomnia vs hypersomnia). You also see (second row) the networks estimated based on the 3 multiple indicator methods. Again, we don’t know which of these methods got it “right” because we don’t know the true structure, but we can compare how they perform, and how similar they are. For this, we use a simulation study I will not describe in detail here.
What conclusions can we draw from this empirical study in the STARD data (there is also a simulation study on this dataset which I skipped here for the sake of brevity)?
When the sample size is small, different single-indicator networks (i.e., those based on a single questionnaire) produced more distinct results, as would be expected in the context of measurement error. As sample size increases, agreement between single-indicator networks grows substantially. Consistent with the simulation results [of the prior study], each of the methods for incorporating multiple indicators are in reasonably close agreement with each other.
Conclusions
For our main conclusion, I’ll just quote from the paper.
We found that, when measurement error was present, incorporating multiple indicators per variable considerably improved the performance of network models. Networks based on average scores, factor scores, and latent variables each demonstrated comparably good sensitivity and a strong correlation between edges in the estimated and true networks for samples as small as 500. For sample sizes greater than 1,000, latent network models outperformed both average scores and factor scores, exhibiting both better precision and better specificity. Thus, while large samples may mitigate the effects of measurement error, these simulation results suggest that it is considerably more efficient to address measurement error by improving node reliability (e.g., by gathering multiple indicators per node).
In the remainder of the manuscript, we discuss the role of measurement error in the next decade of the network psychometric literature, and how mitigate potential issues that arise from measurement error.
PS: The paper was a collaboration led by Jill de Ron, but I wrote this blog post, which means that all errors and mistakes here are mine only.